Overview
Research Focus
Select Publications
Facilities
Projects
Research Staff
Overview
Cancer biology
Cancer continues to be the leading cause of mortality worldwide and contributes significantly to the socio-economic issues across all societies and nations. Understanding and developing treatments for cancer is a challenge for the scientific community across the globe. Research at the Cell and Molecular Biology laboratory is primarily focused on understanding the molecular mechanisms underlying tumor metastasis with particular emphasis on the role of cell junction complexes during tumor progression. Studies are also focused on understanding the genotypic and phenotypic characteristics of normal cancer cells and cancer stem cells, the unique features that can be utilized as criteria/markers for rapid and early detection of cancers. Other areas of research interest include understanding the cancer genome heterogeneity among the subpopulation of cancer cells in tumor using computational tools and to elucidate the mechanism of anti-cancerous properties of specific medicinal plants.
Ion Channel Biology
Ion channels are a group of transmembrane proteins that form the basis of the electrochemical properties of living cells. They help charged molecules to cross the lipid bilayer. The ion channels are responsible for the electrical properties of nervous and cardiac systems. From transmission of nerve impulse to the maintenance of calcium homeostasis, ion channels play a central role in the regulation of body functions. Numerous studies indicate the role of ion channels in various pathophysiological conditions, making them interesting drug targets. Our team uses various techniques of molecular biology, protein chemistry, fluorescence imaging and patch clamp physiology to understand the role of ion channels in the physiology and pathophysiology of nervous system. The unit focuses on the function and regulation of pannexin channels and their role in pain physiology with the objective being to develop drugs specifically targeting these channels.
Research Focus
Cancer biology
Dynamics of cell junction proteins during tumor progression
Cell junction complexes play a major role in cell adhesion and migration which are highly regulated during crucial processes like embryogenesis, wound healing and also in perturbed conditions as in tumorigenesis. It is well-established that alterations in cell adhesion processes are the hallmark of aggressive malignant tumors. Much of the research in the cancer over the last decade has been focused on understanding the metastatic potential of cancer cells. Metastasis is a complex multi-step process involving cell invasion, migration, intra/extravasation, and changes in cell adhesion properties. Once the tumors are formed at the primary site, they shed some of the cells to the circulatory system through a process termed as Epithelial to Mesenchymal transition (EMT).These circulating tumor cells (CTC) disseminate into distant sites and colonize, through a reverse process to EMT called Mesenchymal to Epithelial transition (MET). Both EMT and MET helps cells to re-establish at target site, thus completing the metastatic loop as shown in figure.

Schematic representation of the metastasis formation by circulating tumor cells (CTCs) undergoing epithelial to mesenchymal transition (EMT) and mesenchymal to epithelial transition (MET) changes in their cellular characteristics.
The circulating tumour cells (CTC) are different from the cells at primary tumour in that they have metastatic potential. Some of the specific clonal population of CTC called cancer stem cells (CSC) are responsible for driving tumour recurrence and metastasis. These CSC have the capacity of self-renewal and have long term proliferative potential and are known to be highly resistant to chemo and radiotherapy. Since metastasis is the main reason for the tumour associated deaths, understanding the molecular mechanism of CTCs biology and the EMT process are the key focus of current research. During EMT, the cancer cells change their adhesive property to more of migratory and invasive characteristics which also involve dramatic reorganization of cytoskeleton and the cell junction complexes. The molecular processes that determine such cellular changes are poorly understood and many dynamic cell junction proteins require better functional and molecular characterization. In order to better understand the molecular mechanism of regulation of cell junction proteins, it is relevant to identify the potential binding partners of these cell junction proteins. To identify the potential binding partners, a global approach of screening the cDNA library of humans was undertaken using yeast two hybrid system. A schematic representation of identifying protein-protein interaction using human cDNA library is shown in the figure 2 (A and B) and the representative positive clones of the library screening is shown in the figure.

(A) Schematic representation to determine protein-protein interaction through yeast two-hybrid system. The protein of interest “X” is expressed as a fusion of GAL4-DNA binding domain and forms a bait plasmid and the protein “Y” is expressed as a fusion of GAL4 activation domain and this represents the prey plasmid. If both the proteins interact with each other, their interaction drives the expression of reporter genes.
(B) Schematics of the Yeast two hybrid library screening of one of the cell junction protein along with the human cDNA library.
(C) Representative of the diploids growing on selective nutrient media. The blue colonies represent the interaction between two proteins.
Phenotypic and Genotypic characterization of different subpopulations of cancer cells
Recent studies have shown that CSC exhibit genotypic and phenotypic characteristics that are different from normal cancer cells. The differences observed include size, density, electric charges, deformability, expression of unique cell surface markers and genomic variability. Such vast differences among CSC and other subpopulation of cancer cells can be used as important criteria to detect CSC in early stage cancers using Atomic Force microscopy (AFM). AFM has emerged as a multifaceted tool that can differentiate cellular variations of single living cells at nano scale in their natural environment. AFM provides an opportunity to visualize the minute details of cellular cytoskeleton and genomic variabilities with high resolution imaging system. Cancer cells exhibit remarkable changes in viscoelastic properties, cell stiffness morphology, nuclear and chromatin architecture, an important criteria to be considered for AFM studies. Studies in our lab are focused on understanding the structural and morphological changes that differentiates CSC from other subpopulation of cancer cells using Atomic force microscopy (AFM). The data obtained from AFM studies will be carefully analyzed and mathematical cell models will be developed. Depending on the specificities and sensitivity of these developed models, these studies can be utilized as non-invasive methods for cancer diagnosis. Schematic representation of the cellular difference among different subpopulations of cancer cells are detected by AFM studies.
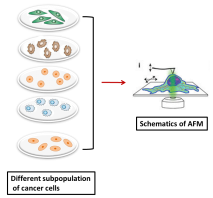
Schematic view of Atomic force Microscopy (AFM) method. AFM probe is aligned over cytoplasm, nuclei and nucleoli of the cells to calculate elastic moduli by force –indentation curves.
Cancer Genomics
Recent studies and evidences have supported the theory that cancer is the disease of genome and cancer cells exhibit high genomic heterogeneity within the tumour. Whole genome sequences of single cancer cells within the tumour using next generation sequences (NGS) technologies have enabled us to understand the clonal evolution of mutation and how these mutations have contributed to tumour progression and therapeutic resistance. To better understand the mutational status in cancers, we along with our collaborators in the computational engineering laboratory are developing databases for identifying multiple cancer biomarkers from whole genome sequencing data. The developed databases can be utilized for diagnosis, clinical and translational research and for academic purposes. The information obtained from NGS data holds great promise for personalized cancer therapy.
Ion Channel Biology
Patch Clamp Technique:

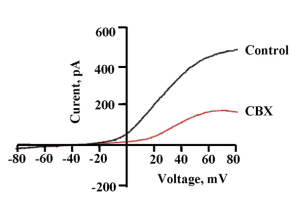
‘Patch clamp’ technique is widely used to study hemichannel. In this method current and voltage changes occurring across a cell membrane can be recorded directly. For voltage gated channels like Panx1, voltage clamp recording is used. In voltage clamp, the membrane potential is clamped at a desired voltage and the resulting changes in current are recorded. Usually the voltage is varied from hyperpolarizing to positive potentials either stepwise (step protocol) or as gradient (voltage ramp protocol). This gives an I-V curve that tells about the voltage dependent behaviour of the channel. Patch clamp recording can be done in two ways: single channel recording and whole cell recording. Single channel recording gives the unitary conductance of the channel and tells about the different sub-conductance states involved. The open probability and gating of the hemichannel can be analysed. In whole cell recording, ensemble activity of the channels is recorded.

Whole cell patch clamp technique
(http://www.science-display.com/new/datenbank/detail.php?action=edit&id=23)
Studying the role of Pannexinhemichannels in purinergic signalling.
Extracellular nucleosides such as adenosine and nucleotides like ATP act as danger signals by binding to a special class of membrane receptors known as purinergic receptors. Danger signals act as a sign if tissues are distressed. One of the major pathways of ATP release is through the activated hemichannels. Panx1 hemichannel is the major ATP release channel. Through the release of ATP, hemichannels participate in purinergic signalling. Panx1 plays a central role in purinergic signalling in various physiological events like taste sensation, vascular tone regulation, immune cell activation, inflammation etc. ATP is added extracellularly, it can increase membrane permeability. Molecules as large as 1 kDa may diffuse across ATP-permeabilized plasma membrane. It is believed that the opening of hemichannels and P2X receptors are involved in the ATP-induced membrane permeabilization. The mechanism of large pore formation by P2X7R is not well understood. There is a controversy whether the pore is formed by P2X7R itself that dilates over time or is it a completely unique entity activated downstream of P2X7R (North, 2002). There are evidences supporting both the theories. Our lab focuses on investigating the interaction between P2X7 receptor and Pannexin 1 hemichannels in membrane pore formation.

Schematic representation of pathways in membrane pore formation
Pharmacological characterization of Pannexin 1 hemichannels.
Panx1 plays a major role in the immune system, where it is involved in the innate immune response and cell death, inflammation, and the neutrophil activation which involves Panx1 mediated ATP release. Panx1 channel activity is implicated in various diseases, including AIDS, epilepsy, Crohn’s disease and oxygen glucose deprivation in neurons. Recent reports have suggested that Panx1 is required for metastasis of tumor cells. While in another study Panx1 was found to have tumor suppressive effects in glioma cells. Being a major player in pathological signalling, Panx1 is a potential target for therapeutic intervention in many diseases.
There is a lot of confusion in the literature about Panx1 function and behaviour. Lack of selective inhibitors has made the study of Panx1 even more challenging. We are working towards identification of highly selective modulators of Panx1. Although a number of drugs are able to inhibit Panx1 channels, the search for a compound that is specific only with this protein continues.
Post translational regulation of Pannexin 1 hemichannels.
Panx1 forms a non-selective large pore in the cell membrane, allowing passage of large molecules up to molecular weight of 1 kDa. The opening of the channels need to be regulated strictly. Otherwise, prolonged opening will cause the leakage of cellular constituents, affecting the homeostasis and finally cell death. Regulation of a channel protein can happen at three different levels such as post translational modification, Regulated trafficking of the protein and Regulation of the opening and closing of the channel. Regulation at post translational level can happen through glycosylation, nitrosylation and phosphorylation. The phosphorylation of Panx1 has not been studied in detail. Though few indirect studies imply phosphorylation of the protein, but not extensively reported. The team is interested to thoroughly investigate the phosphorylation of Panx1 which will lead to the understanding of regulation of Panx1 and its mechanism.
Select Publications
Nitric oxide inhibits the pannexin 1 channel through a cGMP–PKG dependent pathway. Poornima V., SirishaVallabhaneni, MohonaMukhopadhyay, Amal Kanti Bera. Nitric Oxide (2015) 47: 77–84. IF:3.548
Crosslinking biopolymers for biomedical application. Narendra Reddy, Roopa Reddy, Qiuran Jiang. Trends in Biotechnology (2015) 33(6):362–69. IF:11.126
A Review on Protein Functionalized Carbon Nanotubes. Kathyayini Nagaraju, Roopa Reddy, Narendra Reddy. Journal of Applied Biomaterials and Functional Biomaterials (2015) 18:13(4):e301-12. IF:1.069
3rd.Stratifin (14-3-3 σ) limits plakophilin-3 exchange with the desmosomal plaque. Roberts BJ, Reddy R, Wahl JK PLoS One (2013) 4; 8(10):e77012. IF:2.806
P2X7 Receptor–Pannexin 1 Hemichannel Association: Effect of Extracellular Calcium on Membrane Permeabilization. Poornima V, Madhupriya M, Kootar S, Sujatha G, Arvind Kumar and Amal Kanti Bera J. Mol. Neurosci. (2012) 46(3): 585-94. IF:2.89
Facilities
EVOS XL core Imaging system, Veriti 96 well thermal cycler, BioRad- Chemi Doc XRS+ Imaging system, Thermo Scientific Multiskan GO, Thermo Scientific 1300 class II, B2 Biological safety cabinet, Thermo Scientific BB 150 Co2 incubator, Thermo Scientific Legend micro centrifuge
Projects
- A study on the effect of P2X7receptor on Pannexin 1 function. – DBT
- Development of computational pipeline to identify multiple biomarkers for the diagnosis of cancer and other genetic disorders- SSPS
- Differentiating Cancer stem cells from other subpopulation of cancer cells through Atomic force microscopy (AFM): Implications in the early detection and diagnosis of cancer- SSPS
Research Staff

 Roopa Reddy is a Genetics and Cancer Biology researcher, currently working as an Assistant Professor. Roopa after her initial grooming in India pursued her studies in USA, where she obtained MS in the field of Veterinary and Biomedical Sciences from University of Nebraska (UNL), Lincoln, USA in 2008 and Ph.D in Medical Sciences from University of Nebraska Medical Centre (UNMC) Omaha, USA in the year 2013. With almost nine years of research experience in different domains of biology including Microbiology, Genetics and Molecular biology, Cancer biology and Bioinformatics, her current research is focussed on understanding cell junction complex dynamics during tumour progression. Using yeast as a model organism, she identified many novel protein-protein interactions and the potential binding partners of the protein of interest by screening whole cDNA library of humans. Her research interest is also extended in elucidating the phenotypic and genotypic differences between normal cancer cells and cancer stem cell, which can help in early detection of cancer. With further focus on Cancer genomic, Roopa along with other collaborators from computational engineering group is developing the software for identifying multiple cancer biomarkers from whole genome sequence data of cancer patients, which holds a great promise for personalised cancer therapy. The proposed software can be applicable for diagnosis, clinical and translational research and for academic purposes. Other areas of research interest include understanding the host pathogen interactions in disease conditions and in elucidating the mechanism of action of anti-cancerous properties of selected medicinal plants.Roopa Reddy is also a recipient of prestigious Fellowship from University of Nebraska (UNL), USA during her MS program. In recognition to her work in cancer research during her Ph.D program, she was awarded the prestigious Graduate research Assistant /Fellowship through University of Nebraska Medical Centre (UNMC), USA, for three consecutive years which was awarded on highly competitive basis. To add to her scientific records, she has publications in some of the high impact factor international journals. She has also presented her doctoral research work in several seminar series organised by the host institute and also presented her research work in international conferences organised by the American society of cell biology (ASCB) in annual meeting held at Denver, Colorado.
Roopa Reddy is a Genetics and Cancer Biology researcher, currently working as an Assistant Professor. Roopa after her initial grooming in India pursued her studies in USA, where she obtained MS in the field of Veterinary and Biomedical Sciences from University of Nebraska (UNL), Lincoln, USA in 2008 and Ph.D in Medical Sciences from University of Nebraska Medical Centre (UNMC) Omaha, USA in the year 2013. With almost nine years of research experience in different domains of biology including Microbiology, Genetics and Molecular biology, Cancer biology and Bioinformatics, her current research is focussed on understanding cell junction complex dynamics during tumour progression. Using yeast as a model organism, she identified many novel protein-protein interactions and the potential binding partners of the protein of interest by screening whole cDNA library of humans. Her research interest is also extended in elucidating the phenotypic and genotypic differences between normal cancer cells and cancer stem cell, which can help in early detection of cancer. With further focus on Cancer genomic, Roopa along with other collaborators from computational engineering group is developing the software for identifying multiple cancer biomarkers from whole genome sequence data of cancer patients, which holds a great promise for personalised cancer therapy. The proposed software can be applicable for diagnosis, clinical and translational research and for academic purposes. Other areas of research interest include understanding the host pathogen interactions in disease conditions and in elucidating the mechanism of action of anti-cancerous properties of selected medicinal plants.Roopa Reddy is also a recipient of prestigious Fellowship from University of Nebraska (UNL), USA during her MS program. In recognition to her work in cancer research during her Ph.D program, she was awarded the prestigious Graduate research Assistant /Fellowship through University of Nebraska Medical Centre (UNMC), USA, for three consecutive years which was awarded on highly competitive basis. To add to her scientific records, she has publications in some of the high impact factor international journals. She has also presented her doctoral research work in several seminar series organised by the host institute and also presented her research work in international conferences organised by the American society of cell biology (ASCB) in annual meeting held at Denver, Colorado.
Email Id: roopa.reddy@ciirc.jyothyit.ac.
Poornima V Poornima is presently working as Scientist. She obtained her PhD in ion channel physiology from IIT Madras, Chennai in 2014. She studied BSc Biotechnology from BMS women’s college, Bangalore and MSc Biochemistry from Bangalore University. She was university topper in MSc Biochemistry in the year 2007 and is the recipient of three gold medals. She was awarded summer research fellowship from JNCASR in 2004 and worked in Indian Institute of Science as an intern. She has been an active participant in science and literary clubs during her school and college days. Teaching is her passion and she has taught different courses on biochemistry and physiology to undergraduate and postgraduate students. She has also worked with high school students to introduce creative science teaching and activity – based learning methods. As a passionate researcher, she is interested to decipher the role of ion channels in brain physiology and patho-physiology. Her work on a gap junction protein, Pannexin 1 and a purinergic receptor, P2X7, has received high number of citations in international journals in a very short period of time. Her work was selected for the well known Gordon research conference on ion channels in 2010. She wishes to continue her study on these proteins and develop therapeutic strategies to combat neurological disorders like ischemia, Alzheimer’s and cancers like glioblastoma. She has been awarded DBT- research associateship in 2017. She has published research articles in reputed international journals. Apart from research, she enjoys art and craft, cooking and loves reading fiction.
Poornima is presently working as Scientist. She obtained her PhD in ion channel physiology from IIT Madras, Chennai in 2014. She studied BSc Biotechnology from BMS women’s college, Bangalore and MSc Biochemistry from Bangalore University. She was university topper in MSc Biochemistry in the year 2007 and is the recipient of three gold medals. She was awarded summer research fellowship from JNCASR in 2004 and worked in Indian Institute of Science as an intern. She has been an active participant in science and literary clubs during her school and college days. Teaching is her passion and she has taught different courses on biochemistry and physiology to undergraduate and postgraduate students. She has also worked with high school students to introduce creative science teaching and activity – based learning methods. As a passionate researcher, she is interested to decipher the role of ion channels in brain physiology and patho-physiology. Her work on a gap junction protein, Pannexin 1 and a purinergic receptor, P2X7, has received high number of citations in international journals in a very short period of time. Her work was selected for the well known Gordon research conference on ion channels in 2010. She wishes to continue her study on these proteins and develop therapeutic strategies to combat neurological disorders like ischemia, Alzheimer’s and cancers like glioblastoma. She has been awarded DBT- research associateship in 2017. She has published research articles in reputed international journals. Apart from research, she enjoys art and craft, cooking and loves reading fiction.
Email Id: poornima.v@ciirc.jyothyit.ac.in
