Overview
Computational Engineering is a relatively new discipline that deals with the development and application of computational models and simulations, often coupled with high-performance computing, to solve complex physical problems arising in engineering analysis and design. The research focus of this laboratory is on the use of heterogeneous parallel processing to speed up the bioinformatics applications such as cancer genomics pipelines, data science and analytics, modeling and simulation in medical applications, sensors simulation and simulation of molecular dynamics for studies in computational sciences.A heterogeneous cluster computing facility has been created to meet the huge computational needs of all these research activities. Other ongoing research areas include cyber security, blockchain technology,Internet of Things and development of (i) pulse analyzer device for use in traditional medicine and(ii) bioinformatics toolbox.

Cancer Genomics
Genomics is the study of the genes in the genome and their interactions with the environment. Genomic analysis concerns withbiomolecules such as DNA sequences and RNA sequences in (i) identifying them,(ii) comparing their features, and (iii) measuring their structural variation, gene expression, or regulatory and functional element annotation. Next Generation Sequencing (NGS) technologies help in the characterization of point mutations of a wide range of cancers and their structural alterations. Complete genome sequences of numerous cancer types are becoming increasingly available, which provide a comprehensive view of cancer development and growth.Cancer genome studiesenable understanding the gene abnormalities that are the root cause of many types of cancer. This improved understanding of cancer biology helps in developing new waysof diagnosis and treatment.
Cancer accounts for approximately 13% of all deaths worldwide and contributes significantly to global socio-economic burden to the society. There exists evidenceto support the theory that cancer is a disease of genome; aberrant genomic alterations are the hallmark of cancer cells. At the computational engineering lab, current research in cancer genomics is focused mainly on understanding intratumor heterogeneity whichplays a major role in tumor metastasis and drug resistance to therapeutics. To better understand the tumor genome heterogeneity in tumor niche, a global approach of sequencing the whole cancer genome was undertaken. Since then, several sequencing studies have identified a strong correlation between genomic alterations with tumor progression and resistance to conventional therapies. However, effective treatment of cancer rely on early detection, precise diagnosis, and implementation of specific therapeutic strategies that give rise to the concept of personalized medicine.
Although, whole genomic sequences provide important insights into clonal evolution of mutations, lack of proper tools and pipelines to analyze and interpret the framework of data has limited the use of sequencing technology for diagnosis and therapeutics. Independent studies have identified several driver mutations with specific cancer types, but analyzing how each of these mutations work individually or in a cohort towards tumor progression is limited. Recent advancement in computational biology and mathematical modeling has shown great potential in analyzing huge sequencing data and in understanding the origin and progression of cancer without much of the wet lab experiments and ethical issues.

Figure 1: Genome Analysis Pipeline
Genome Analysis Pipeline
A computational model for identifying multiple cancer specific biomarkers from the whole genome sequence data of cancer patient and analyzing cancer prognosis using predictive machine learning models is proposed. The source of the whole genome sequencing data of cancer cells will be obtained from the European Genome-Phenome Archive (EGA), The Cancer Genomics Hub (CGHub) and other relevant databases. The source for the potential biomarkers known for each cancer type is will be obtained from 1000 genomes browser and COSMIC databases and the reference sequences of these biomarkers will be obtained from NationalCenter for Biotechnology Information (NCBI). The pipeline developed will be a cluster of four main groups based on their application such as assembly, alignment, variation discovery and annotation. For predictive modeling, machine learning approaches will be used, that have unique characteristics for prediction and classification such ascapacity control of the decision function, the use of the kernel functions, and the scarcity of the solution. The three-way data split using this model will be applied for training, validation, and testing. The proposed pipeline and a schematic representation of the frontend of the pipeline are shown below.
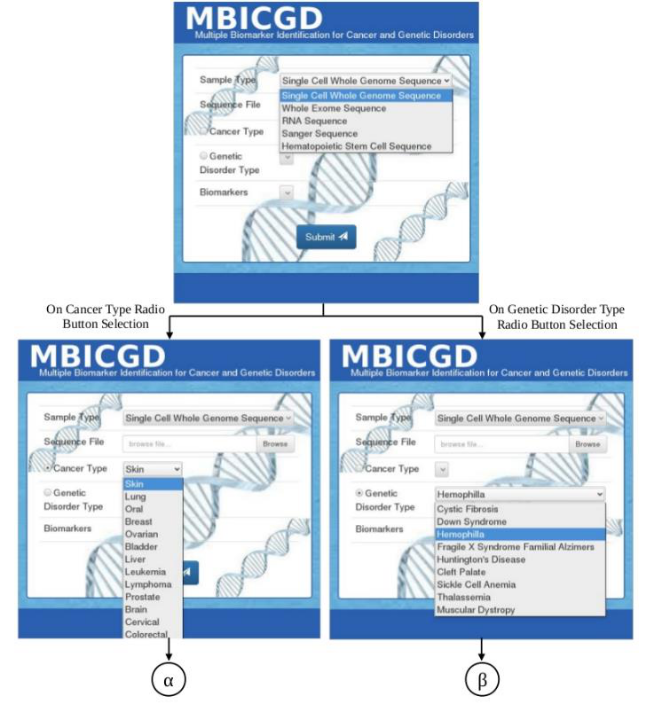
Figure 2(a): Frontend of the MBICGD Pipeline

Figure 2(b): Frontend of the MBICGD Pipeline (contd.)
Research Objectives
- To recognize and use new opportunities offered by NGS technologies in the field of cancer genomics
- To overcome challenges raised by the massive amount of sequence data that has been and will be generated
- To create a dedicated bioinformatics facility for genome analysis related to cancer studies
- To empower biomedical teams working in the field of cancer to best exploit the huge data generated by the ongoing genomic projects
Research Outcome
- High-throughput tools for DNA and RNA characterization that facilitate comprehensive analyses of cancer genomes including all somatic alterations.
- Cancer genome methodology to understand the relationship between the genetic mutations and the clinical response to assist in cancer therapy.
- New methodologies for cancer diagnosis and prognosis.
- Comprehensive catalogs of the genomic changes in respect of specific cancer types.
- Technologies for complete molecular profiling of tumors enabling genome-informed personalized cancer medicine.
- New techniques for validation of genomic data to (i) distinguish mutations responsible for disease pathogenesis resulting from genomic instability, (ii) define genes responsible for cancer initiation, progression, and maintenance, and (iii) identify the effective ways to therapeutically exploit this information.
Beneficiaries
- Cancer patients
- Medical practitioners
- Genome scientists
- Research scholars
Wrist Pulse Analyzer
Wrist pulse analysis is a simple non-invasive technique used in ancient Indian medicine Ayurveda and traditional Chinese medicine (TCM)to diagnose health. Most modern pathological diagnosis tools give reports in terms of content and chemical changes in the patient’s body. Very few devices give reports based on body constitution of the patient. Wrist pulse diagnosis is an ancient method of diagnosing human body constitution. Computerized pulse diagnosis uses sensors to acquire wrist pulse signals and uses machine learning techniques to analyze patient’s health based on the acquired pulse signals.Wrist pulse indicates the significantly varied blood flow of an organ in abnormal health conditions. Patient health conditions manifest themselves in reflected waves of the wrist pulse signals in different ways. With careful interpretation of these signals, one can perform an initial diagnosis to great accuracy.
Research Objectives
- To design an efficient and accurate pulse acquiring device without losing vital radial pulse information of the patient
- To acquire radial arterial acoustic vibrations through a non-invasive pulse detector
- To develop an indigenous application to analyze, characterize and classify the human radial pulse obtained through the detector
- To provide low-cost pulse diagnosis system for alternative and integrated medical practitioners
To provide complete analysis of the overall constitution of a person in accordance with the TCM and Ayurveda principles through charts and bar graphs

Figure 3: Wrist Pulse Analysis
Research Outcome
- A wrist pulse detector which takes acoustic vibrations of the radial arterial pulse and an intelligent machine which analyses these variations and provides vital five elements energy level information which aids in the correct diagnosis of ailments.
- Low-cost pulse diagnosis system for alternative and integrated medical practitioners.
- A system for complete analysis of the overall constitution of a person in accordance with the TCM and Ayurveda principles through charts and bar graphs.
Beneficiaries
- Alternative medicine practitioners (AYUSH) for getting complete body constitution of the patient.
- Patients in terms of getting better and affordable alternative therapies.
- Researchers to pursue their research on pulse diagnosis and alternate therapies.
Whole-Cell Modeling and Simulation
Understanding and engineering of biological systems require comprehensive models of cellular physiology with 100% predictability. These whole-cell models guide experiments in molecular biologyand enable simulation and computer-aided design in synthetic biology. Theyhelp personalized medical treatment. Constructing comprehensive whole cell models with sufficient detail and validating them is a massive work. Modeling larger cells and more complex physiology involve (i) model building and integration, (ii) model validation, (iii) data curation, (iv) experimental interrogation, (v) analysis and visualization, (vi) accelerated computation, (vii) community development and (viii) collaboration. A broader multidisciplinary research community is needed to innovate in all these areas. As a first step, a study on the issues and challenges is initiated at the lab and is in progress.

Figure 4: Whole-Cell Modeling Process
Research Objectives
- To set up and configure tools for modelling.
- To estimate parameters and feed the estimated data to a tool.
- To design submodules depending on the cell process.
- To design a model by combining all the submodules.
- To speed up the whole-cell model simulations on high-performance heterogeneous platform, exploiting parallel computing technology.
Research Outcome
- The precise model by making use of available whole-cell model software and tools.
- To represent cell functions and characteristic using whole-cell modelling.
- To validate the new model with experimental data.
Beneficiaries
- Clinicians
- Bioinformatics scientists
- Research scholars
Cyber Security
Digital technology is growing rapidly, causing a wide spread of distribution of digital documents and images over the Internet. The security of digital documents, images and other multimedia data has thus become extremely important for common people and the government. Secured transmission of digital images is an important issue in information security field. Cryptographic encryption techniques provide an effective security to data by converting it into an un-understandable form to attackers. Conventional cryptographic encryption methods are unsuitable for image encryption. In recent years, chaotic theory has attracted research community for image encryption. A simple and secured scheme for image encryption using one-dimensional logistic maps has been studied. This image encryption scheme first shuffles the position of pixel values and then changes the gray values leading to a complex relationship between the original plain image and the encrypted image. Two operations, namely, image scrambling and diffusing, are performed by logistic maps. Various experiments test the robustness and the security aspects of the algorithm and it is found that such schemes are resistant to different cryptanalytic attacks and provides adequate security.

a) Original Image b) Encrypted Image
Figure 5: Image Encryption based on Chaotic Theory
Tongue Diagnosis using Deep Learning
Analysis of the tongue is crucial in the evaluation of the human body in Ayurveda medicine. There is substantial research scope for tongue diagnosis in Ayurvedic therapy. Tongue pictures are classified into vata, pitta and kapha (tri-dosha) with the color, coating, texture and shape features. These characteristics would also be determined largely by the portion of the tongue part (upper region, middle region and sides). Segmentation is the most important procedure in defining the region of interest in the tongue image. These features include various color models and differentiated parts of the tongue body. Coating and background are also considered for feature set to feed them to the training-testing model for classification. The final outcome of the research is to blend in with the traditional Ayurvedic methods to the machine learning and deep learning model as it describes the basic class of inspection in medication.

Figure 6: Tongue Diagnosis using Deep Learning
- S Balaji, Roopa Reddy et.al. (2019), “Recent Advancements in Machine Learning and Artificial Intelligence Techniques for Cancer Diagnosis”, International Research Journal of Engineering &Technology, 6 (5), May 2019, pp 4599-4613, IF: 7.34
- Nayana G. Bhat, S. Balaji (2019), “Whole-Cell Modelling and Simulation: A Brief Survey”, New Generation Computing, 38 (1), pp 259-281, DOI 10.1007/s00354-019-00066-y, IF: 0.83
- Sathish P.K., S.Balaji (2019), “Fusion of Image Feature Descriptors for Person Re-identification”, International Journal of Engineering and Advanced Technology, 9 (2), Dec. 2019, pp 4993-4998, DOI: 10.35940/ijeat.B2700.129219, IF: 5.97
- Harshvardhan Tiwari, Virtual machine selection optimization using nature inspired algorithms, International Journal of Mathematics and Computer Science, ISSN 1814-0432, Vol. 16, pp. 677-685, 2021 IF: 0.83.
- Harshvardhan Tiwari, Virtual Machine placement Using Energy Efficient particle swarm Optimization in cloud data center, Cybernetics and information technologies, 1314-4081, Vol. 21, pp. 62 – 72, 2021 IF: 1.74.
- Harshvardhan Tiwari , Merkle-Damgard Construction Method and Alternatives: A Review, Journal of Information and Organizational Sciences ISSN 1846- 3312,Vol. 41, pp. 283-304, 2017. IF: 0.93.
- Pallavi C.V., S.Usha, S.Balaji (2020), Harshvardhan Tiwari, “IoT Architecture for Agriculture Applications: A Formal Assessment”, International Journal of Advanced Science and Technology, 29 (6), 2020, pp. 7974-7986, IF: 0.41.
- Sumanth N .S., N. Satish Kumar, Harshvardhan Tiwari, Balaji S., Prabhanjan S., Meenakshi Malhothra, Pallavi C.V. (2020), “Application of Machine Learning Techniques for Tongue Diagnosis in Ayurveda”, Journal of Advancements in Robotics, 7 (1), 2020, pp 8-14, IF 6.05
- Kurubara Basavaraj, S.Balaji (2021), “Nadi Pariksha: A Novel Machine Learning Based Wrist Pulse Analysis through Pulse Auscultation System Using KNN Classifier”, International Journal of Electronics and Communication Engineering and Technology, 12 93), Sep– Dec 2021, pp 1-10.
The primary mission of the Computational Engineering Laboratory (CEL) is the delivery and support of a High-PerformanceComputing (HPC) resource to be used by faculty members, research scholars and students. Seven servers, ten workstations and two NAS systems are connected using 10Gbps interconnect switch in a cluster. The HPC setup currently has 110 CPU Cores and 1168 GB RAM with a theoretical TFLOPS rating of 62.71664 TFLOPS. It is a highly scalable setup and is targeted to reach around 1000 cores in near future.

- “Testing and Measurement of Homoeopathic Medicines Potency Using Their Colligative Properties” sponsored by Central Council for Research in Homeopathy, Ministry of AYUSH, GoI.
- “Digitalization of Best-before Date of the Packaged Meals Using a Smart Label (SCENSIOR)” sponsored by Global Innovation & Technology Alliance, a Public Private Partnership (PPP) between Technology Development Board (TDB), DST, GoI, and India’ s apex industry association, Confederation of Indian Industry (CII). DST-GITA.
- Artificial Intelligense powered gradation equipment for lemon.
- “Establishment of a Genome Computing Cluster”, KFIST-I Level, KSTePS, Vision Group of Science and Technology (VGST), GoK.
- Fruit Crops Yield Estimation using Machine Vision Techniques, VTU
- Machine and deep learning based disease prediction model, Genbioca Science Pvt Ltd.,
- Coffee plant disease identification using CNN, Aroma Coffee Works
- “Whole-cell Modeling with accelerated Simulation on Heterogeneous Parallel Platform”, Research Grant for Scientists/Faculty (RGS/F),KSTePS, Vision Group of Science and Technology (VGST), GoK.
- “Development of Computational Model for Identification of Multiple Biomarkers in Whole Cancer Genome and Predictive Model for Cancer Prognosis” (SSPS)
- “Wrist Pulse Analyzer”
- “PanchNidaan”

Balaji
Balaji received his Ph.D. in Computer Science and Engineering from Indian Institute of Science, Bengaluru in 1993. His thesis,S-NETS: A Tool for the Performance Evaluation of Hard Real-Time Scheduling Algorithms, is on hard real-time systems. His registration for M.Sc.(Engg.) in Fault-Tolerant Computing in the same department was upgraded in 1989 to Ph.D. due to the outstanding progress made at graduate level. He holds a Master’s Degree in Applied Mathematics from Anna University (1981) and a B.Sc. in Mathematics, Physics and Statistics from Madras University (1979).
Balaji is a technologist turned academician with a blend of academic and industrial experience, and expertise in successfully executing and managing R&D intensive projects. He has considerable experience in developing mission and safety critical applications and cutting-edge systems in embedded and mobile computing. He also brings vast experience at senior management levels during his stint in ISRO Satellite Centre, the country’s premier space research organization. His forte is in ensuring that all the functional units within an organization are aligned to deliver best value to the organization. His strong technical foundation and his experience at senior management levels help him move seamlessly between finance, administration, project management and engineering. He has defined and established software processes for developing mission and safety-critical space applications. He has architectedseveral simulation packages for the design and design validation of attitude and orbit control systems of satellites. He has introduced numerous innovations for the design validation of spacecraft onboard sub-systems and developed advanced test systems to validate satellite control systems. He setup the facility for software development using high-level programming languages ADA, processor-based satellite control systems, institutionalized processes and methodologies for the development of such systems and has trained/mentored staff to deliver high integrity systems that meet critical spacecraft mission requirements. He has developed several in-house tools for software engineering and project management.
He has served at AMC Engineering College and City Engineering College in various capacities, such as Professor, Vice-principal and Principal. He, then, worked at the Centre for Emerging Technologies, Jain University, where he initiated research in the areas of bioinformatics, genomics and molecular modelling and simulation.
Balaji is a Fellow of IETE and a Senior Member of IEEE. He is a also member of ISTE, Astronautical Society of India, Association for Computing Machinery, and Institute of Smart Structures and Systems. His current research interests include heterogeneous parallel processing, data science and analytics, bioinformatics, genomics, computational sciences, modeling and simulation, video data mining and analytics, image processing, embedded and real-time computing, IoT, and mobile computing.
During his tenure at ISRO Satellite Centre, Balaji was (i) a member/convener of several software review committees, (ii) a member/convener of several software verification and validation committees, (iii) a member of Seminar Committee, (iv) a member of Library Committee, (v) Executive Secretary, Editorial Board, Journal of Spacecraft Technology, and (vi) Executive Secretary, Association for Autonomous and Fault-Tolerant Systems. He was the Organizing Secretary of the Information Technology Track of the Indian Science Congress, 2002.
Balaji has delivered invited talks on recent trends in computer science and engineering at various national conferences and workshops. He was a member of the Technical Committee of a few national conferences. He has been a member of the Board of Studies in Computer Science and Engineering in a couple of engineering colleges. He served as the internal representative of all Research Review Committees – Computer Science and Engineering, Jain University, from March 2014 – Jul 2016. He has been a member of the Comprehensive Examination Committees of VTU and has been a thesis examiner at Anna University. He is an Authorized Research Supervisor in Computer Science and Engineering at VTU, Jain University and Tumkur University. Nine scholars have successfully completed their Ph.D. degrees under his guidance. He has published over fifty research articles in refereed national and international journals. He has authored chapters in three books published by Springer and CRC Press. He is currently a Professor at the Centre.
Email Id: balaji.s@ciirc.jyothyit.ac.in

Harshvardhan Tiwari
Harshvardhan Tiwari received his Ph.D. degree in CSE from JIIT University, Noida, UP, India. He completed his Prost-Graduation (M.Tech.) and graduate study (B.E.) both in Computer Science and Engineering from RGTU, Bhopal, MP, in 2009 and 2005 respectively. His research interests are Machine Learning, Data Science, Deep Learning, Computer Networks, Algorithms, DBMS, and Cryptography. He has more than 9 years of teaching and research experience. He has several publications to his credit that are SCOPUS and DBLP indexed. During his academic career, he taught courses such as Python Application Programming, Machine Learning, Operating System, Computer Architecture and Organization, Database Management System, Cryptography, Computer Networks, and Web Technologies. Apart from teaching, he is an active researcher and currently an advisor of 4 research scholars. He is also a member of the technical program committee of referred journals and conference proceedings. He is actively involved in curriculum planning and syllabus design at an engineering college for graduate level program. He has been an external examiner at other universities. He has participated in different FDPs and is a member of many professional scientific bodies such as IEI, IAENG, ICST.
Email Id: harshvardhan.t@ciirc.jyothyit.ac.in

Nayana G Bhat
Nayana G Bhat received her Bachelor’s degree in EEE from VTU and Master’s in CSE from Kuvempu University. Currently, she is her Ph.D from VTU. She has taught several undergraduatecomputer science courses and has guided a number of undergraduate projects. She has more than 8 years of teaching experience and is a researcher having 4+ years of experience in whole-cell modelling and simulation. She has participated in many national and international conferences, FDPs, seminars and workshops. She is a member ofCSI and ISTE. Her research interests include bioinformatics, parallel computing, and image processing. She is currently an Assistant Professor at the Centre.
Email Id: nayana.gb@ciirc.jyothyit.ac.in

Sripadaraja K
SripadarajaK completed his Master’s in electronics from Bangalore University and has submitted his doctoral thesis at NIT, Surathkal. He started his career in MEMS as a project assistant at the Indian Institute of Science. Later, he joined Bigtec in 2009 as an Applications Engineer. He was a Senior Application Engineer of the IntelliSense software team that set up a new company, SriDutt Technologies Pvt. Ltd., and further worked as a Product Manager. He has conducted several hands-on training programs and has given many invited talks across India. He has published many papers and won research grants. Currently, at CIIRC, he is involved in the development of MEMS biosensors for various diagnostics applications. He is a consultant and a researcher having 12+ years of experience in MEMS sensors/actuators design and development.

Akhilesh S
Akhilesh S holds a Bachelor’s degree and Master’s degree, both in CSE, from VTU. He has participated in several national and international conferences and FDPs. His research interests include AI and ML, computer vision, and blockchain technology.

Chitrabalan M
Chitrabalan M received his Bachelor’s degree in EEE from Sri Manakula Vinayagar Engineering College, Pondicherry University. He has participated in many national and international conferences, and FDPs. His research interests include embedded systems, mobile computing, and blockchain technology. He is currently a Research Fellow at the Centre.

Ranjith Kumar K
Ranjith Kumar K has a Bachelor’s degree in ECE from REC College, Anna University. He has participated in many national and international conferences, FDPs, and seminars and workshops. His research interests include embedded programming and graphics animation. He is currently a Research Fellow at the Centre.
